240: 低酸素誘導因子(Hypoxia-Inducible Factors)
酸素は不可欠なもので、それがないと私たちの細胞はたちまち死んでしまう。このため酸素の量を監視し、低酸素症(hypoxia)と呼ばれる酸素が少ない状態になれば応答することに特化したしくみを発達させてきた。酸素が不足した細胞は、多くの赤血球を生み出しより多くの血管をつくるよう身体に伝える信号を送り出す。また、代謝のしくみを変えて、あまり多くの酸素を必要としないエネルギー代謝経路を使うようにする。例えば、ピルビン酸脱水素酵素(pyruvate dehydrogenase)を減らし、乳酸脱水素酵素(lactate dehydrogenase)を増やすというようにして。この年のノーベル生理学・医学賞は、HIFシステム(低酸素誘導因子システム、hypoxia-inducible factor)と呼ばれるこの中心的な酸素検知過程のしくみに関する分子的な詳細を明らかにした3人の研究者に贈られた。
酸素が十分あるとき
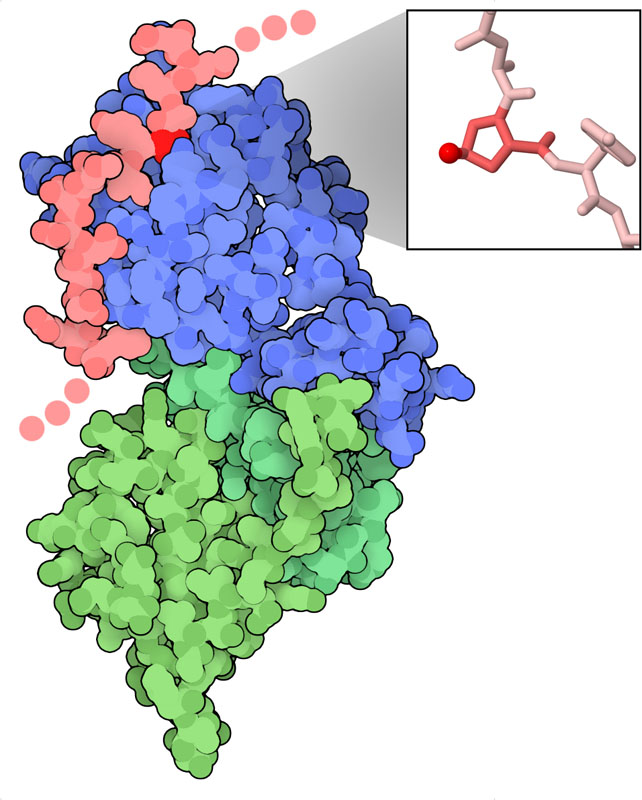
低酸素誘導因子α(hypoxia-inducible factor、HIF-α)は、細胞が酸素に制約がある状態に応答するしくみの中心となるスイッチである。約800個のアミノ酸からできた、複数の機能部位で構成されるタンパク質である。ここに示す構造(PDBエントリー1lqb、1lm8)には配列中央の小さな一部分が含まれ、ここには2つの重要なプロリン残基がある(ここにはそのうちの一方を示している)。酸素が豊富にあるとき、このプロリンはHIFプロリンヒドロキシル化酵素(HIF prolyl hydroxylases、PHD酵素)によってヒドロキシル化される。そしてこのヒドロキシプロリン(hydroxyproline)は、HIF-αをユビキチン化とプロテアソームによる分解対象となるようにするpVHL(フォンヒッペル・リンドウ病腫瘍抑制タンパク質、von Hippel-Lindau disease tumor suppressor)を含んだ複合体によって認識される。そのため、通常の酸素濃度であれば、HIF-αは常時抑制されており、細胞は通常通り活動する。
酸素の検知
PHD酵素は酸素濃度を検知する働きを担っている。金属イオンと補助基質のα-ケトグルタル酸(α-ketoglutarate)を使い、HIF-αにあるこの二つの重要なプロリンに酸素原子を付加する。酸素が不足すると、PHDの触媒作用は抑えられプロリンは修飾されなくなる。またFIH(HIF抑制因子、factor inhibiting HIF)と呼ばれる別の酵素が、HIF-α中にあるアスパラギン酸をヒドロキシル化して転写機構(PDBエントリー1h2n、ここには示していない)と相互作用する方法変える第2のヒドロキシル化反応を行う。
酸素が不足したとき
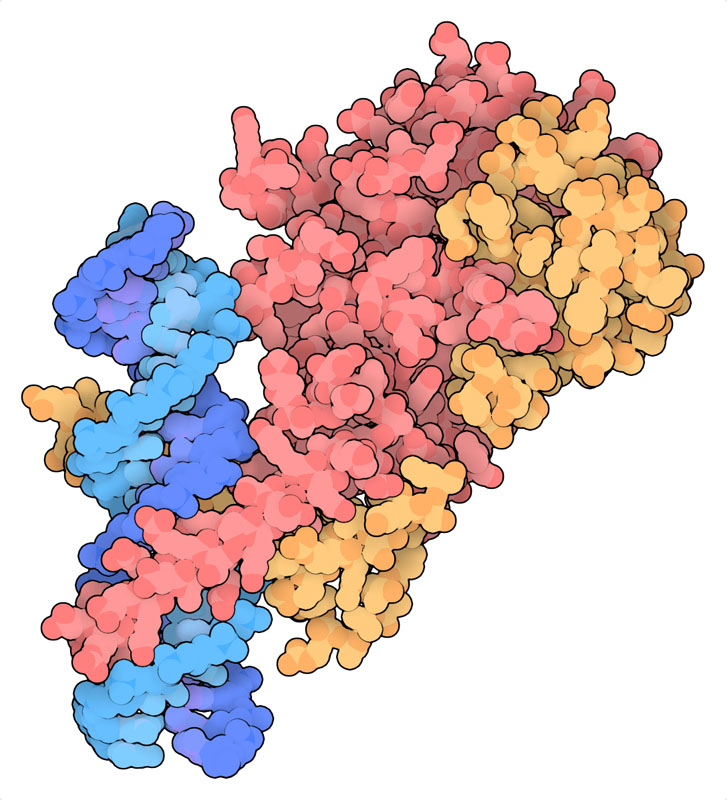
酸素濃度が低くなると、HIF-αはヒドロキシル化されずプロテアソームに分解されることもなくなって活動を始める。核へと移動してHIF-βと呼ばれる仲間のタンパク質と連携し、ゲノムのさまざまな部位と結合する。それにより低酸素代謝に関する遺伝子の転写を促し、酸素がより良く行き渡るよう呼吸系の体制を変更する。ここに示す構造(PDBエントリー4zpr)には複合体のDNA結合部位がDNAの短い断片と結合したものが含まれている。
構造をみる
対話的操作のできるページに切り替えるには図の下のボタンをクリックしてください。読み込みが始まらない時は図をクリックしてみてください。
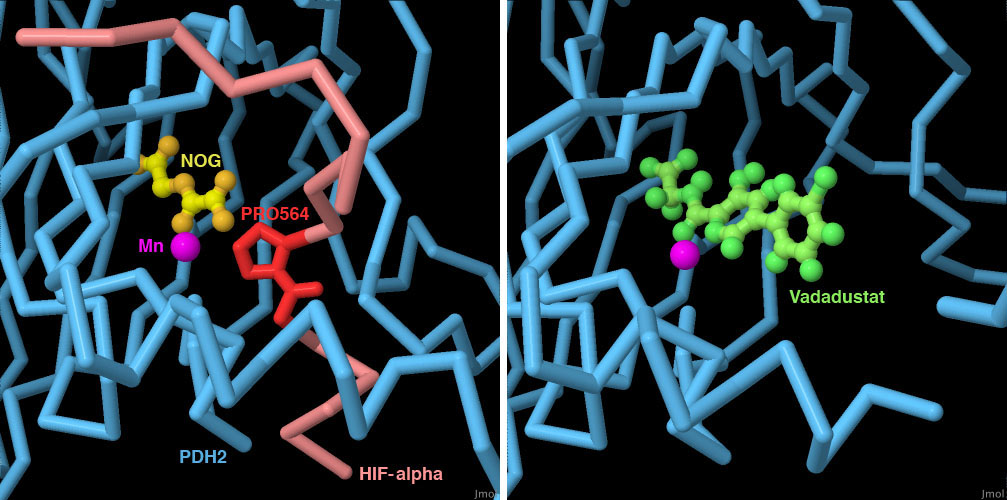
PHD酵素に結合する薬は貧血症(anemia)の治療薬として適当なのかについて査定が行われているところである。この種の薬はPHD酵素の働きを阻害することにより、細胞にもっと酸素が必要だと思わされ、より多くの赤血球をつくるための信号を送らせる。PHD2の初期状態での構造(PDBエントリー2g1m、2g19)があることで、構造に基づく酵素阻害剤の設計が可能となった。ここに示すのは現在評価中の薬バダデュスタット(Vadadustat、PDBエントリー5ox6)である。この構造とをHIF-αの小さな断片と結合したPHD2の構造(PDBエントリー3hqr)と比較することにより、薬がα-ケトグルタル酸が酵素とつくる結合をまねていて、HIF-αのプロリンへの結合を妨げるのに十分な大きさがあることを見ることができる。なお、3hqrに含まれるNOGはα-ケトグルタル酸に似た化合物である。
理解を深めるためのトピックス
- PDBにはFIH(HIF阻害因子)の構造がたくさん登録されていて、どんな構造をしているのかや、基質や阻害剤とどのように相互作用するのかを見ることができる。
- HIF複合体にはハサミの形をした「塩基性らせんループらせん」(basic helix-loop-helix、bHLH)と呼ばれるDNA結合ドメインがある。「bHLH」をPDBで検索すると他の例も見ることができるだろう。
参考文献
- 5ox6 2017 Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials. Chem Sci 8 7651-7668
- 4zpr 2015 Structural integration in hypoxia-inducible factors. Nature 524 303-308
- 2012 Hypoxia-inducible factors in physiology and medicine. Cell 148 399-408
- 3hqr 2009 Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure 17 981-989
- 2g1m、2g19 2006 Cellular oxygen sensing: crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2). Proc.Natl.Acad.Sci.USA 103 9814-9819
- 1h2n 2003 Structure of factor-inhibiting hypoxia-inducible factor (Hif) reveals mechanism of oxidative modification of Hif-1Alpha. J.Biol.Chem. 278 1802-1806
- 1lqb 2002 Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 417 975-978
 の生体高分子学習ポータルサイト
の生体高分子学習ポータルサイト