162: Dermcidin
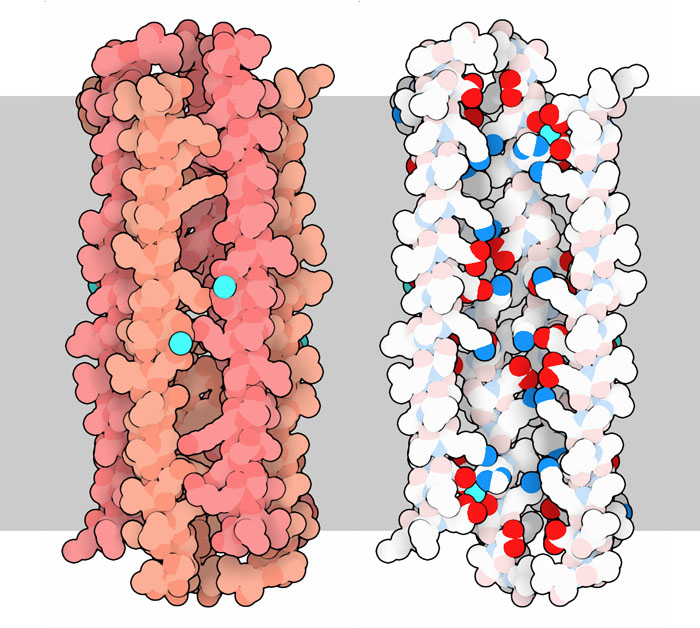
Small toxic peptides are used by our cells in the battle against bacterial infection.
For more details, please refer the original article in PDB-101 of RCSB PDB.

Small toxic peptides are used by our cells in the battle against bacterial infection.
For more details, please refer the original article in PDB-101 of RCSB PDB.
This is an mirror page of the "Molecule of the Month" article provided in PDB-101 of the RCSB PDB. This article was released on June 2013. To reprinting and citation, please refer our terms and conditions page.


VR / 3D glasses & game


Molecular viewer with VR (Virtual Reality)
Web service viewing biological molecule structures for learning biology with enjoy!

Making protein molecules with paper

Encyclopedia of Protein Structures including the explanation of biopolymer in PDB

Resources of previous workshops and events

Pelmanism and snake game
Explanation about COVID-19

PDBj
[Protein Data Bank Japan]
{
"header": {
"minimamHeightScale": 1.0,
"scalingAnimSec": 0.3
},
"src": {
"spacer": "/share/im/ui_spacer.png",
"dummy": "/share/im/ui_dummy.png"
},
"spacer": "/share/im/ui_spacer.png"
}